

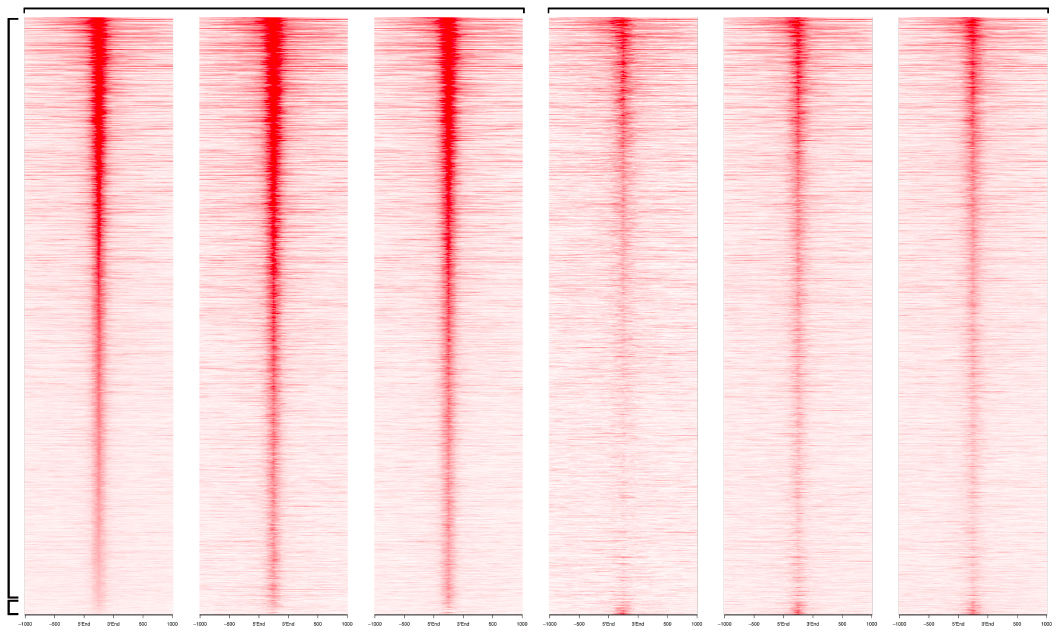
Although in vitro studies have implicated genome reorganisation as a central mechanism coordinating cellular differentiation and cell fate determination during cell differentiation, it has not been possible to study long-range enhancer-promoter reorganisation at a genome-wide level during cell differentiation in vivo. This project brings together a group that has developed novel methods to isolate primary neuronal progenitors from the developing mouse brain along a defined developmental trajectory in vivo (Basson) with a group that has and continues to develop innovative molecular and bioinformatics approaches to study chromatin organisation and promoter-enhancer interactions in the genome (Osborne).