
Synthetic community [SynCom] transfer for the...
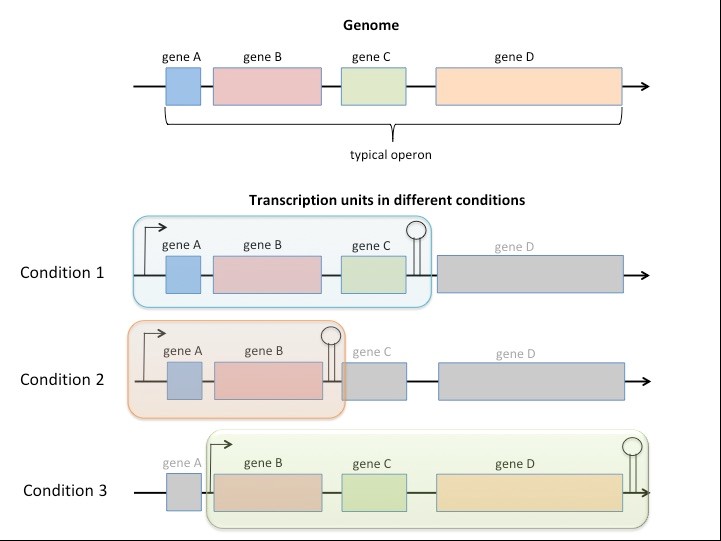
Advances in the methods profiling transcriptomes have offered new insights into bacterial operons. What were once thought to be relatively simple and stable units of transcription have been shown to be surprisingly dynamic in response to environmental challenges experienced by bacteria. In this project we will harness the power of third generation (long-read) sequencing and computational biology methods to analyse operon variability in mycobacterial genomes and investigate its regulation by protein and non-coding RNAs