

It is widely viewed that EIEC and Shigella species evolved from E. coli via independent acquisitions of a large virulence plasmid (pINV) encoding a type 3 secretion system (T3SS). Sequence Type (ST)99 O96:H19 E. coli is a novel clone of EIEC responsible for recent outbreaks in Europe and South America.
Here, we use 92 whole genome sequences to reconstruct a dated phylogeny of ST99 E. coli, revealing distinct phylogenomic clusters of pINV-positive and -negative isolates. To study the impact of pINV acquisition on the virulence of this clone, we developed an EIEC-zebrafish infection model showing that virulence of ST99 EIEC is thermoregulated. Strikingly, zebrafish infection using a T3SS-deficient ST99 EIEC strain and the oldest available pINV-negative isolate reveals a separate, temperature-independent mechanism of virulence, indicating that ST99 non-EIEC strains were virulent before pINV acquisition. Taken together, these results suggest that an already pathogenic E. coli acquired pINV and that virulence of ST99 isolates became thermoregulated once pINV was acquired.
Enteroinvasive Escherichia coli (EIEC) and Shigella are etiological agents of bacillary dysentery. Sequence Type (ST)99 is a clone of EIEC hypothesized to cause human disease by the recent acquisition of pINV, a large plasmid encoding a type 3 secretion system (T3SS) that confers the ability to invade human cells. Using Bayesian analysis and zebrafish larvae infection, we show that the virulence of ST99 EIEC isolates is highly dependent on temperature, while T3SS-deficient isolates encode a separate temperature-independent mechanism of virulence. These results indicate that ST99 non-EIEC isolates may have been virulent before pINV acquisition and highlight an important role of pINV acquisition in the dispersal of ST99 EIEC in humans, allowing wider dissemination across Europe and South America.
Enteroinvasive E. coli (EIEC) and Shigella species are Gram-negative, human-adapted pathogens that cause bacillary dysentery. The greatest burden of bacillary dysentery is in low- and middle-income countries (LMICs) (1), although the true burden of EIEC infection is likely underestimated since it is difficult to distinguish from Shigella. Historically, Shigella was classified as its own genus, with four distinct species, but Multi-Locus Sequence Typing (MLST) and whole-genome sequencing data clearly show Shigella spp. are lineages of E. coli, as are EIEC (2, 3). Each Shigella and EIEC lineage evolved independently within the E. coli population, following the horizontal acquisition of a ~220 kbp virulence plasmid (also known as plasmid of invasion or pINV) from a currently unknown source (2). pINV encodes a type three secretion system (T3SS) that facilitates the invasion of human epithelial cells and is thermoregulated in both EIEC and Shigella (4).
A novel clone of EIEC, of serotype O96:H19 and Multi-Locus Sequence Type (ST) 99, was first described in 2012 in Italy and has since caused several foodborne outbreaks of moderate to severe diarrheal disease across Europe and South America (5-7). Before 2012, ST99 E. coli had not been reported in the literature as causing human disease but had been sporadically isolated from cattle and environmental sources (8). ST99 EIEC isolates have been characterized as possessing the virulence hallmarks of EIEC and Shigella (pINV and T3SS) (9), but its metabolic capacity closely resembles that of commensal E. coli and it has more recently been associated with pga-mediated biofilm formation (6, 9). It has therefore been proposed that ST99 EIEC diverged recently from ST99 E. coli due to the acquisition of pINV.
The zebrafish (Danio rerio) larvae model is widely used to study infection biology in vivo because of its rapid development and innate immune system that is highly homologous to that of humans (10, 11). Zebrafish have emerged as a valuable vertebrate model to study human enteropathogens like Shigella (12), highlighting the key roles of bacterial virulence factors (e.g., T3SS and O-antigen) (13, 14) and cell-autonomous immunity (e.g., autophagy and septin-mediated immunity) (12, 15) in host-pathogen interactions.
In this observation, we reconstruct a dated phylogeny of ST99 E. coli using publicly available whole genome sequences, to understand the role of pINV in its global dispersal. We develop a temperature-dependent zebrafish infection model to assess the virulence of EIEC and non-EIEC ST99 isolates, highlighting the power of zebrafish infection in studying the evolution of novel enteropathogens causing disease in humans.
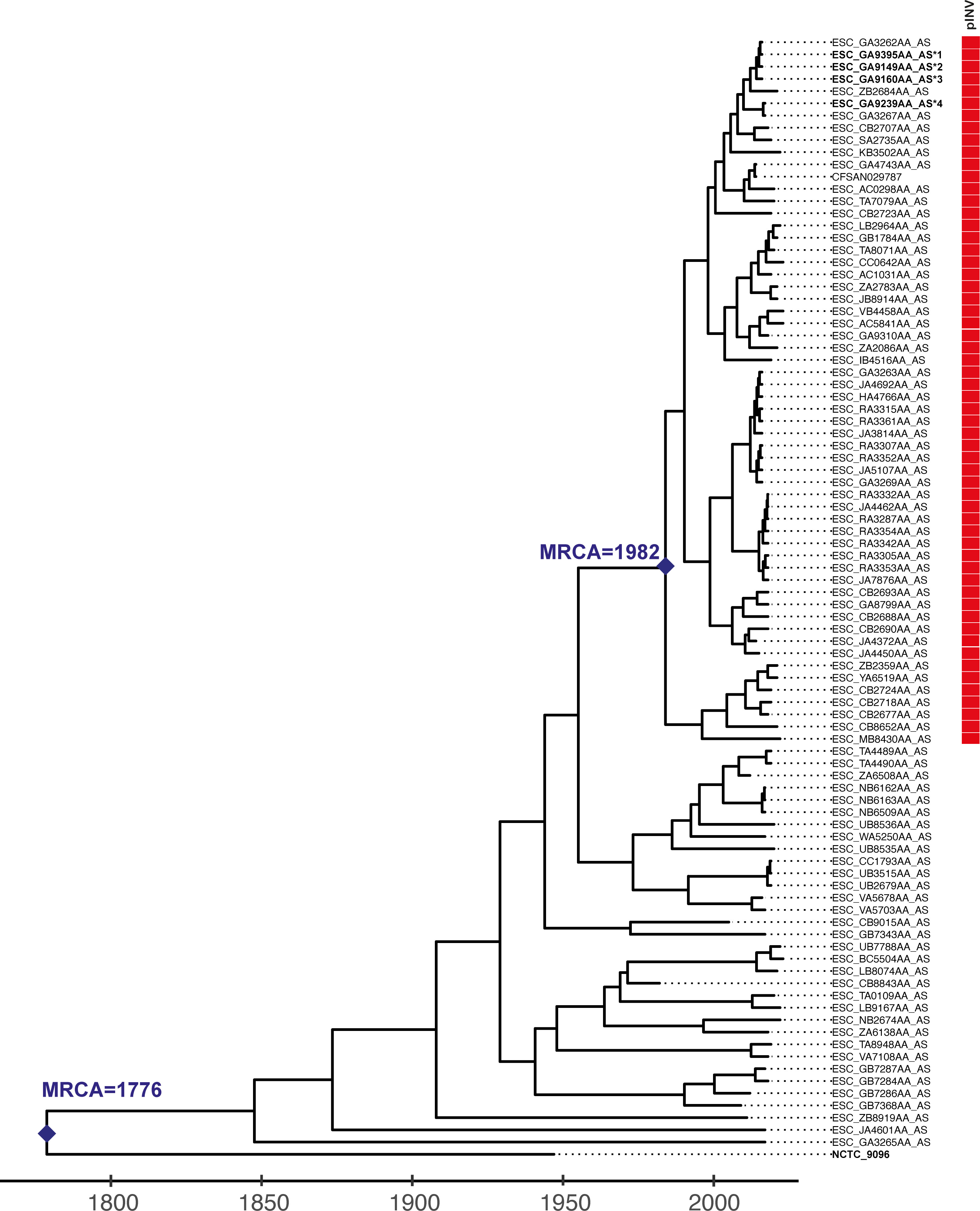
To dissect the evolution of the ST99 clone and its transition to EIEC, we analyzed all publicly available ST99 genomes (n = 92), using the EnteroBase integrated software environment (16). EnteroBase routinely scans short-read archives and retrieves E. coli and Shigella sequences from the public domain or uses user-uploaded short reads. We used Gubbins v.3.2.1 (17) to filter recombinant sites, RaxML v.8.10 to infer a Maximum Likelihood phylogenetic tree and BactDating v.1.2 (18) to date the phylogeny (Fig. 1), as previously described by Didelot and Parkhill (19). Root-to-tip genetic distances were positively associated with the year of isolation (R2 = 0.19, P = 6 × 10-3), and the date-randomization test showed no overlap between results of observed and date-randomized analyses (Fig. S1), indicating a moderate molecular clock signal to support dating analysis. From this analysis, we estimate that the most recent common ancestor (MRCA) of the whole ST99 group (pINV+ and pINV–) existed circa 1776 [95% highest posterior density (HPD), 1360–1927]. To test for the presence of pINV, we used ShigEiFinder, which scans the genomes for pINV-encoded genes and deems an isolate positive for pINV when 26 of 38 genes are present (20). The pINV+ isolates form a distinct cluster, with their MRCA existing circa 1982 (95% HPD, 1965–2011) (Fig. 1). This suggests that the ST99 EIEC may have been circulating undetected for ~30 years before being detected in the 2012 outbreak.