
Synthetic community [SynCom] transfer for the...
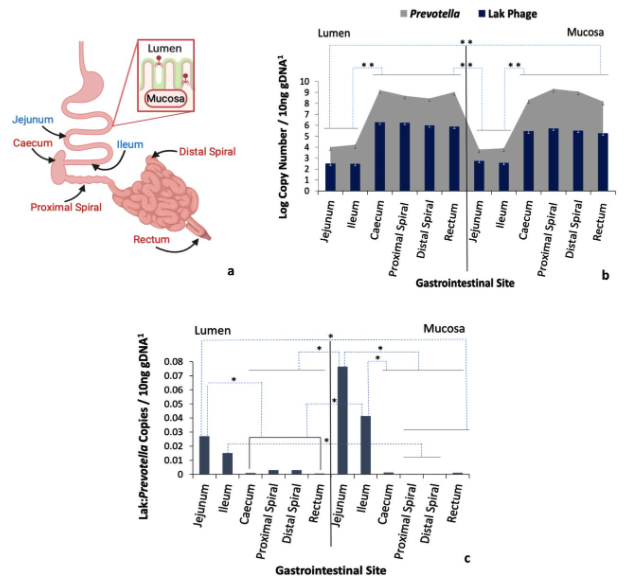
Lak phages were detected in 13 different animal types and are particularly prevalent in pigs, with significant enrichment in the hindgut compared to foregut. We reconstructed 34 new Lak genomes, including six curated complete genomes, all of which are alternatively coded. The most deeply branched Lak is from a horse faecal sample and is the largest phage genome from an animal microbiome (~660 kbp).
From the Lak genomes, we identified families of hypothetical proteins associated with specific animal types. Overall, we substantially expanded Lak phage diversity and demonstrate their occurrence in a variety of human and animal microbiomes.