

Indeed, if you watched a bunch of cells growing in a dish for a couple of days, you’d notice that over time each clone exhibits differences in its division profile. Some cells will divide (much) faster than others and populate (much) more progeny but why these differences occur and how such cell cycle diversity is maintained remains unclear.
To tackle this problem, we developed a double whammy of tools to follow cell lineages over several generations. We implemented our fully automated time-lapse microscopy movie acquisition hardware only to collect live-cell imaging data, which we analysed using our custom-developed fully automated machine (deep) learning and computer vision toolkit. These data helped us to shine some light onto how single cells behave & what drives their cell cycle towards a particular duration. And your guess is correct, the analysis was, again, fully automated.
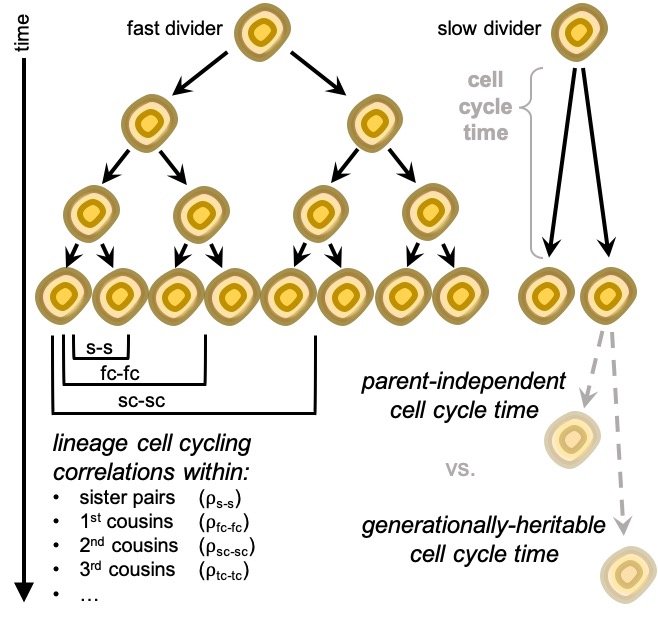
We collected a unique evidence based on analysis of more than 20,000 single-cells organised in 5,000+ lineage trees which points towards cell cycling duration being a generationally heritable property. To confirm this, we looked at extended family relationships between cells & found that cell cycling remains moderately correlated up to the 4th cousin distance. Cool huh? These cells shared an ancestor more than 7 generations ago, yet remain surprisingly similar in the way and speed they undergo and time their own cell division.
There remains plenty to precisely answer in the upcoming years - is it the cell size, local neighbourhood or growth environment which determine the cell cycling dynamics? But our proprietary software, including a nucleus segmentation network, state labelling classifier, single-cell Bayesian tracking tool & data visualisation plugin, places our research team in a great position to start addressing these burning, single-cell issues.