

Osteoclasts are typically cultured in vitro on a variety of physiological (e.g. cortical bone slices, dentine discs) or non-physiological (e.g. calcium phosphate-coated plates, tissue culture plastic or glass) substrates for analysis of cellular physiology, morphology, and biochemical endpoints. Typical osteoclast parameters measured include tartrate resistant acid phosphatase (TRAP) positivity, number, and resorptive activity as well as multinuclearity (≥ 2 nuclei per cell) and actin ring/ruffled border formation [1,2,3,4]. Of these, number and resorption area provide valuable data about osteoclast formation and activity and have historically been manually quantified through image-processing softwares such as ImageJ [1].
Whilst this method enables user confirmation of individual osteoclasts and associated resorption events, it is time consuming, labour intensive and results in substantial intra- and inter-user variability. Thus, there is a clear need to develop an automated method that allows quick, easy, and accurate analysis of in vitro osteoclast cultures.
Attempts to automate in vitro endpoint analyses have been described but often rely on independent and sequential steps of (1) counting osteoclasts; (2) clearing cells from dentine/bone discs [1, 5,6,7]; and (3) separate measurement of the resorption area [8, 9]. These processes are time consuming and effectively destroy the experiment, preventing revisitation later (e.g. for imaging). Currently, the only attempt to simultaneously quantify osteoclasts and bone surface erosion has been performed on histological sections [10]. TrapHisto is an open-source software integrated into ImageJ that semi-automates histomorphometric analysis of static and dynamic bone turnover parameters, particularly resorption analysis [10].
Recent advances mean that new technologies such as machine learning (ML) can now be used to develop an automated workflow for in vitro osteoclast cultures. ML is an application of artificial intelligence (AI) where constructed mathematical models automatically learn from existing data to create an algorithm that produces accurate predictions from new observations without being explicitly programmed [11, 12]. Supervised ML, such as decision tree algorithms and random forest, requires labelled examples from training datasets. The algorithm learns from the labelled objects and generates a predictive model that accurately sorts new data objects into categories [11, 13, 14].
Application of ML methods has improved understanding and analysis efficiency of complex biological data and processes, especially in genomics, systems biology, and image analysis [11, 15, 16]. However, extensive computational and mathematical knowledge has historically been required to build such ML models, making their application to niche biological questions and processes difficult. The development of ilastik, a free, open-source supervised ML-based bio-image analysis software, has since enabled non-computationally proficient researchers to develop methodologies to rapidly execute complicated image analyses [17]. This user-friendly software contains pre-defined workflows that are adapted by the operator to create bespoke image analysis pipelines whilst completely shielding users from the mathematical and computational complexities required to build the random forest algorithm [14, 17, 18]. Some applications of ilastik include measuring neuronal nuclei and cell bodies and osteoblast differentiation from mesenchymal stem cells [19, 20].
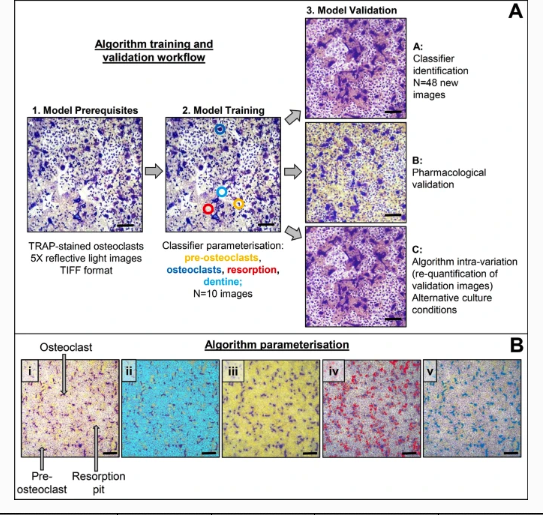
Historically, automatically quantifying osteoclasts in vitro has proven challenging due to the non-uniform cell shape, size, and considerable spacing between nuclei and the cytoplasm of single osteoclasts [8, 21]. Four recent reports have built complex AI-based models to quantify TRAP+ or fluorescently labelled osteoclasts cultured on plastic but not bone or dentine [22,23,24,25]. Resorption parameters were not quantified in any of these models [22,23,24,25]. To date, ML, specifically ilastik, has not been applied to simultaneously measure osteoclast culture endpoints such as osteoclast number and resorption area for cells grown on physiologically relevant substrates. Therefore, the aim of this study was to develop and validate an automated image segmentation workflow in ilastik to reliably, and robustly quantify osteoclast number and resorption area in vitro.